Generate starting locations for activity centers
initialize_classic.RdGenerate a matrix of initial starting locations, possibly accounting for habitat mask.
initialize_classic(y, M, X, ext, site, hab_mask = FALSE, all_random = FALSE)Arguments
- y
Either a matrix or array of encounter history data, possibly from
sim_classic().- M
An integer of the total augmented population size (i.e., detected and augmented individuals). UTMs. An array is used when traps are clustered over a survey area.
- X
Either a matrix or array representing the coordinates of traps in UTMs. An array is used when traps are clustered over a survey area.
- ext
An
Extentobject from therasterpackage. This is returned fromgrid_classic.- site
Either
NULL(if a 2D trap array is used) or a vector of integers denoting which trap array an individual (either detected or augmented) belongs to. Note thatsiteis provided fromsim_classicwhen a 3D trap array is used. However, thissitevariable must be correctly augmented based on the total augmented population size (i.e.,M).- hab_mask
Either
FALSE(the default) or a matrix or array output frommask_polygonormask_rasterfunctions.- all_random
Logical. If
TRUE, then encounter datayare ignored and all initial activity center starting locations are randomly chosen. IfFALSE(the default), then initial values will be the mean capture location for detected individuals and random locations for augmented individuals.
Value
A matrix of initial activity center coordinates with M rows
and 2 columns.
Details
This function generates initial activity center locations based on encounter histories, augmented population size, state-space buffer, and potentially a habitat mask. Note that mean trap detection locations are used for detected individuals while intial values are randomly drawn for augemented individuals. Also, a habitat check will be conducted for all locations when a habitat mask is included.
Examples
# simulate a single trap array with random positional noise
x <- seq(-800, 800, length.out = 5)
y <- seq(-800, 800, length.out = 5)
traps <- as.matrix(expand.grid(x = x, y = y))
# add some random noise to locations
traps <- traps + runif(prod(dim(traps)),-20,20)
mysigma = 300 # simulate sigma of 300 m
mycrs = 32608 # EPSG for WGS 84 / UTM zone 8N
# Create grid and extent
Grid = grid_classic(X = traps, crs_ = mycrs, buff = 3*mysigma, res = 100)
# simulate SCR data
data3d = sim_classic(X = traps, ext = Grid$ext, crs_ = mycrs,
sigma_ = mysigma, prop_sex = 1, N = 200, K = 4, base_encounter = 0.25,
enc_dist = "binomial", hab_mask = FALSE, setSeed = 50)
# generate initial activity center coordinates for 2D trap array without
# habitat mask
s.st3d = initialize_classic(y=data3d$y, M=500, X=traps, ext = Grid$ext,
hab_mask = FALSE, all_random = FALSE)
# make simple plot
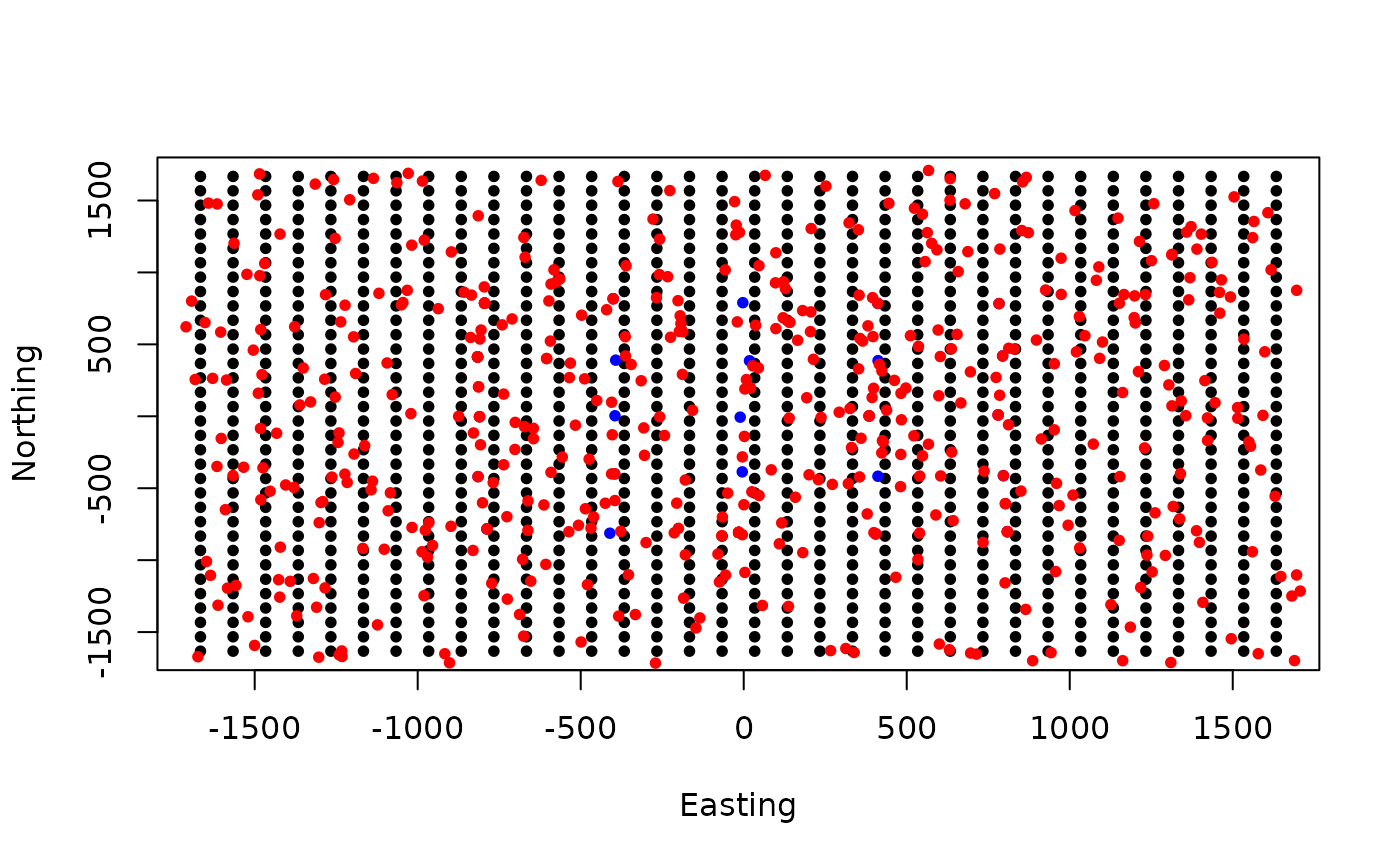
par(mfrow=c(1,1))
plot(Grid$grid, pch=20,ylab="Northing",xlab="Easting")
points(traps, col="blue",pch=20)
points(s.st3d, col="red",pch=20)